Feature extraction
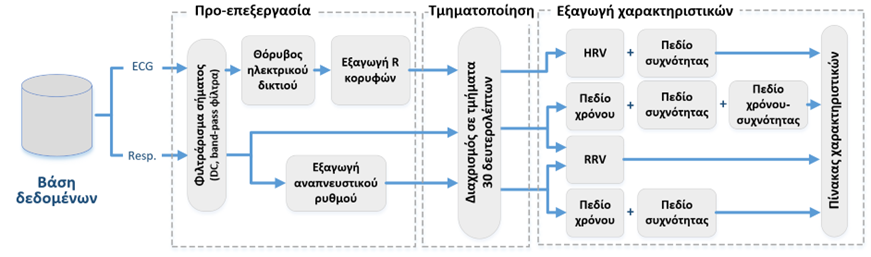
Extracting features from biosignals (e.g. heart/brain electrical activity, breathing, actigraphy, etc.) consists of three main distinct stages of analysis and processing. Initially, the recordings are pre-processed and filtered to minimize the effects of noise and external interference, which usually affects such recordings altering their morphological characteristics. Then, biosignals are typically segmented into smaller intervals of interest, depending on the application to be studied (usually seconds to minutes). This step is particularly important if temporal changes in signal behavior and characteristics need to be considered within the total duration of the recording. Finally, the main stage of the actual feature extraction can be implemented. A wide range of features in the time domain, frequency domain and time-frequency domain can be calculated (depending on the application), along with more specialized metrics such as those from Graph theory, or measures based on heart and respiratory rate variability. An example of the complete cardiac and respiratory activity feature extraction model is shown in the figure below.
Tsiouris, Κ. Μ., Pezoulas, V. C., Zervakis, M., Konitsiotis, S., Koutsouris, D. D., & Fotiadis, D. I., “A long short-term memory deep learning network for the prediction of epileptic seizures using EEG signals,” Computers in biology and medicine, Vol. 99, pp. 24-37, 2018.